Mohammad Dehghanmanshadi
Reducing Domain Gap with Diffusion-Based Domain Adaptation for Cell Counting
Dec 12, 2025Abstract:Generating realistic synthetic microscopy images is critical for training deep learning models in label-scarce environments, such as cell counting with many cells per image. However, traditional domain adaptation methods often struggle to bridge the domain gap when synthetic images lack the complex textures and visual patterns of real samples. In this work, we adapt the Inversion-Based Style Transfer (InST) framework originally designed for artistic style transfer to biomedical microscopy images. Our method combines latent-space Adaptive Instance Normalization with stochastic inversion in a diffusion model to transfer the style from real fluorescence microscopy images to synthetic ones, while weakly preserving content structure. We evaluate the effectiveness of our InST-based synthetic dataset for downstream cell counting by pre-training and fine-tuning EfficientNet-B0 models on various data sources, including real data, hard-coded synthetic data, and the public Cell200-s dataset. Models trained with our InST-synthesized images achieve up to 37\% lower Mean Absolute Error (MAE) compared to models trained on hard-coded synthetic data, and a 52\% reduction in MAE compared to models trained on Cell200-s (from 53.70 to 25.95 MAE). Notably, our approach also outperforms models trained on real data alone (25.95 vs. 27.74 MAE). Further improvements are achieved when combining InST-synthesized data with lightweight domain adaptation techniques such as DACS with CutMix. These findings demonstrate that InST-based style transfer most effectively reduces the domain gap between synthetic and real microscopy data. Our approach offers a scalable path for enhancing cell counting performance while minimizing manual labeling effort. The source code and resources are publicly available at: https://github.com/MohammadDehghan/InST-Microscopy.
Medical Image Segmentation on MRI Images with Missing Modalities: A Review
Mar 11, 2022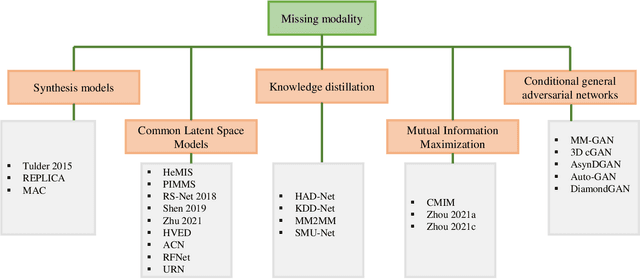
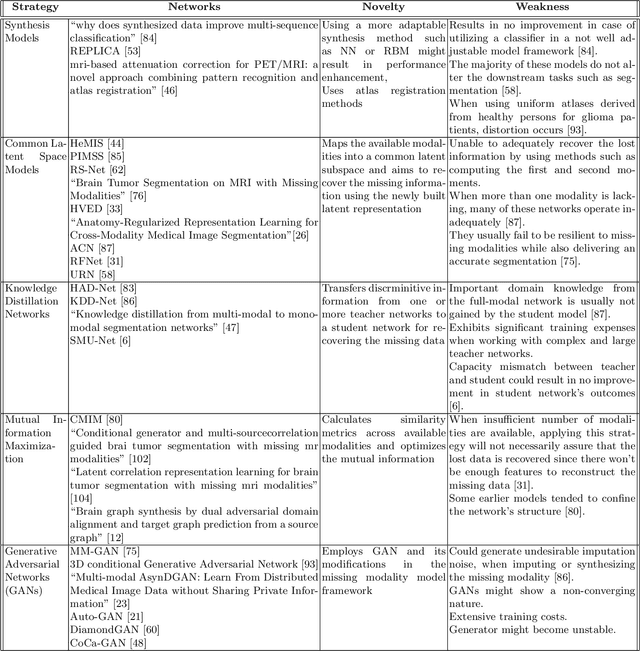
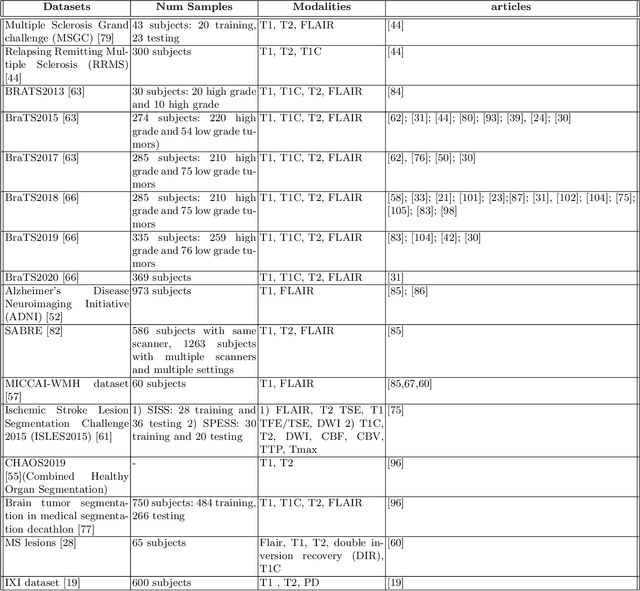
Abstract:Dealing with missing modalities in Magnetic Resonance Imaging (MRI) and overcoming their negative repercussions is considered a hurdle in biomedical imaging. The combination of a specified set of modalities, which is selected depending on the scenario and anatomical part being scanned, will provide medical practitioners with full information about the region of interest in the human body, hence the missing MRI sequences should be reimbursed. The compensation of the adverse impact of losing useful information owing to the lack of one or more modalities is a well-known challenge in the field of computer vision, particularly for medical image processing tasks including tumour segmentation, tissue classification, and image generation. Various approaches have been developed over time to mitigate this problem's negative implications and this literature review goes through a significant number of the networks that seek to do so. The approaches reviewed in this work are reviewed in detail, including earlier techniques such as synthesis methods as well as later approaches that deploy deep learning, such as common latent space models, knowledge distillation networks, mutual information maximization, and generative adversarial networks (GANs). This work discusses the most important approaches that have been offered at the time of this writing, examining the novelty, strength, and weakness of each one. Furthermore, the most commonly used MRI datasets are highlighted and described. The main goal of this research is to offer a performance evaluation of missing modality compensating networks, as well as to outline future strategies for dealing with this issue.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge